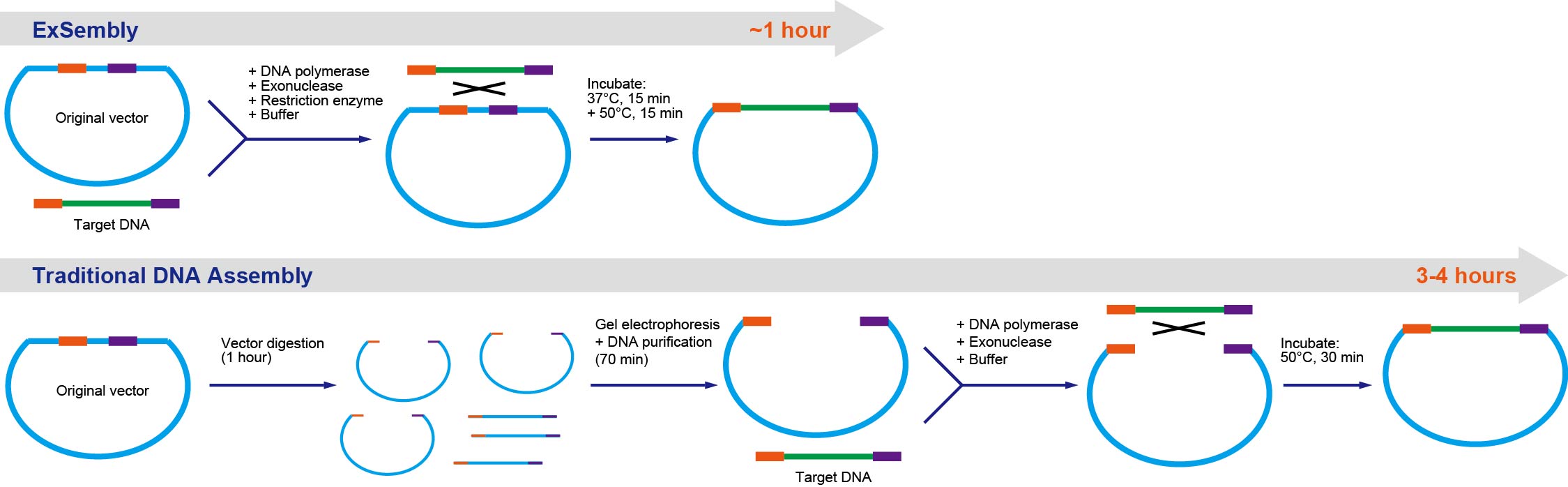
ExSembly™ Cloning Master Mix

ExSembly™ cloning technology enables rapid, single-step, high-efficiency homology-based insertion of any amplified DNA product into a CIRCULAR vector. Vector linearization and DNA fragment insertion occur simultaneously, eliminating time-consuming linear vector preparation steps. Our innovative master mix buffer system supports robust restriction enzyme allowing vector linearization while maintaining high exonuclease and polymerase activities to enable homology-based DNA assembly. Successful fragment insertion destroys the vector-linearizing restriction enzyme site(s). Continuous restriction enzyme activity reduces negative empty-vector clones resulting in high-efficiency (typically >95%) fragment insertion. ExSembly™ cloning mix in assembly cloning kit is compatible with most common restriction enzymes.
Key Features
• One step: combines circular vector linearization and homology-based assembly.
• Rapid: saves 2-3 hours by eliminating vector digestion and gel purification steps.
• Efficient: Typically >95% of colonies bear the correctly inserted DNA fragment.
• Large scale: accommodates up to 500 ng plasmid DNA in one reaction, resulting in more colonies.
• Cost-effective: eliminates vector digestion, gel electrophoresis and purification costs, saving up to 50%.
Customer Testimonials
"I have been using the ExSembly™ Cloning Master Mix for two years and have had great success in cloning a wide range of DNA fragments using this mix. The product is extremely easy to use and makes cloning a very quick process. It is especially ideal to clone multiple DNA fragments simultaneously into a vector. Compared to other approaches and commercial kits I have used in the past; this product's efficiency is very impressive. I highly recommend the ExSembly™ Cloning Mix."
"ExSembly™ Cloning Master Mix works very well in my hands for almost two years. It has similar or higher efficiency compared with other assembly kits!"
Principal
Choose one (or more) restriction enzyme(s*) that linearize the vector and DO NOT cut within the fragment(s) to be inserted. For single fragment insertion, using PCR, add 20 bp homology upstream of the unique restriction site in the vector to the 5’ end of the fragment, and 20 bp of vector homology downstream of the restriction site to the 3' end.
Case Study
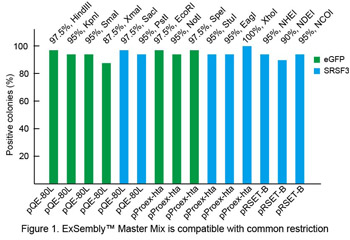 Restriction enzyme compatibility
Restriction enzyme compatibility
ORFs of eGFP or human SRSF3 were PCR-amplified and assembled into vectors (pQE-80L, pProex-hta, pRSET-B) using ExSembly™ Master Mix. ~200 ng vector DNA was used in each ExSembly™reaction. ExSembly™products were purified using a DNA mini-spin column and transformed to DH10B electrocompetent cells and plated on LB/Ampicillin plates. Forty colonies were picked from each reaction to perform colony PCR screening. Percent positive clones are shown in Figure 1 for each of 15 common restriction enzymes.
Comparison between ExSembly™ and Competitor N
Objective:
To compare the efficiency of assembling a single 684 bp PCR product with a 4852 bp vector.
Insert, vector, primers, and restriction enzymes:
A. Gene name: eGFP, 684 bp; Vector: pQE-80L. 4852 bp.
B. Vector: pQE-80L https://www.addgene.org/vector-database/3883/ (4852 base pairs).
C. Forward primer: TCCGCATGCGAGCTCGGTACgcccgccatgaagatcgagt. Upper case bases are nucleotides 148-167 of pQE-80L. Lower case bases are nucleotides 1-20 of the eGFP ORF.
D. Reverse primer: AGTCCAAGCTCAGCTAATTAtcatcgagctcgagatctgg. Upper case bases are the reverse complement of nucleotides 192-211 of pQE-80L. Lower case bases are the reverse complement of nucleotides 665-684 of the eGFP ORF.
E. Restriction enzymes for vector digestion: Kpn I and Hind III
Vector and insert preparation:
A. Vector preparation using agarose gel electrophoresis. 3 μg of pQE-80L DNA was digested with FastDigest Kpn I (FD0524, ThermoFisher) and Fast Hind III (FD0505) for one hour in a 37 °C water bath. The digested DNA was resolved on a 1.0% agarose gel and purified using a standard mini DNA purification spin column protocol. The vector DNA was eluted in 50 μl ultrapure water. The concentration of the vector DNA was adjusted to 30 ng/μl.
B. Vector preparation using DNA spin column. 3 μg of pQE-80L DNA was digested with FastDigest Kpn I (FD0524, ThermoFisher) and Fast Hind III (FD0505) for one hour in a 37 °C water bath. The digested vector was purified using a DNA spin column following a standard protocol and eluted in 50 μl ultrapure water. The eluted DNA was adjusted to 60 ng/μl.
C. Insert DNA preparation. eGFP was PCR-amplified (50 μl) using the primers listed above. The template was pMax-GFP (Kanamycin resistance). The PCR product was purified using a mini DNA spin column, and eluted in 60 μl ultrapure water. Primer dimers were removed after purification using a standard PCR cleanup kit. The concentration of purified PCR product was adjusted to 60 ng/μl.
Experimental set up
| Competitor N Exp. 1 | Competitor N Exp. 2 | ExSembly™ Exp. 1 | ExSembly™ Exp. 2 | |
| Undigested plasmid vector | 0 μl | 0 μl | 2 μl (100 ng/μl ) | 2 μl (300 ng/μl ) |
| Digested plasmid vector | 4 μl (gel purified 30 ng/μl) | 5 μl (column purified 50 ng/μl) | 0 μl | 0 μl |
| Insert | 6 μl | 5 μl | 6 μl | 6 μl |
| KpnI | 0 μl | 0 μl | 1 μl | 1 μl |
| HindIII | 0 μl | 0 μl | 1 μl | 1 μl |
| Water | 0 μl | 3 μl | 0 μl | 0 μl |
| 2× Mater mix | 10 μl | 10 μl | 10 μl | 10 μl |
| Insert: vector | 18:1 | 6:1 | 14:1 | 4.7:1 |
| Incubate temperature and time | 50°C for 45 minutes | 50°C for 60 minutes | 37°C for 15 minutes, and 50°C for 30 minutes | 37°C for 15 minutes, and 50°C for 30 minutes |
Transformation of the assembled DNA to E.coli
The assembled DNA was purified by using a mini DNA spin column and eluted in 50 μl of ultrapure water. 2 μl was used to transform 20 μl DH10B electrocompetent cells. The cells were transferred to 1 ml of SOC medium and incubated at 37 °C with shaking for 1 hour. The cells were then harvested by centrifugation at 7,000 g for 1 minute and plated on LB agar plates containing 100 μg/ml of ampicillin. The plates were incubated at 37 °C overnight.
Colony PCR screening
Colonies were screened by colony PCR using the primers listed above. The positive control was pMax-GFP; no template DNA was used as a negative control.
Results and summary
| Competitor N Exp. 1 | Competitor N Exp. 2 | ExSembly™ Exp. 1 | ExSembly™ Exp. 2 | |
| Vector DNA used (μg) | 3 | 3 | 0.2 | 0.6 |
| Number of colonies | 81 | ~1,200 | ~3,000 | >10,000 |
| Time spent | ~3 hours | ~2 hours | ~1 hour | ~1 hour |
| Positive rate tested | >90% (Not tested)* | 100% (8/8) | 100% (12/12) | >90% (Not tested)* |
* Estimation based on percentage of fluorescent colonies/ total colonies.
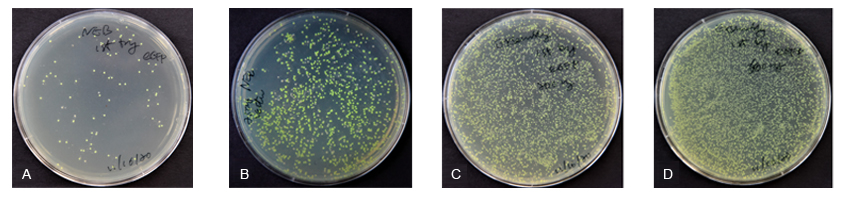
Competitor N Experiment 1: 120 ng gel-purified vector + 360 ng insert (Figure A); Competitor N Experiment 2: 300 ng column purified vector + 300 ng insert (Figure B)
ExSembly Experiment 1: 200 ng vector + 360 ng insert (Figure C); ExSembly Experiment 2: 600 ng vector + 360 ng insert (Figure D)
| Product Name | Cat. # | Size | Price | Order |
| 2× ExSembly™ Cloning Master Mix | M0005 | 10 rxn | $169 |
Related Products
| Product Name | Cat. # | Size | Price | Link |
| 2× LiTaq™ PCR Master Mix | M0024 | 15 ml | $189 | Read more |
| 2× LiSpark™ Ultra SuFi PCR Master Mix | M0031 | 1 ml | $129 | Read more |
| LiPure™ DNA Clean-up Kit | M0011 | 20 rxn | $189 | Read more |
| LiPure™ Plasmid Mini Kit | M0037 | 200 rxn | $189 | Read more |
| LiFect293™ Transfection Reagent | M0002 | 1 ml | $199 | Read more |



